Visualizing Tabular Data
Last updated on 2026-01-13 | Edit this page
Estimated time: 50 minutes
Overview
Questions
- “How can I visualize tabular data in Python?”
- “How can I group several plots together?”
Objectives
- “Plot simple graphs from data.”
- “Plot multiple graphs in a single figure.”
Visualizing data
The mathematician Richard Hamming once said, “The purpose of
computing is insight, not numbers,” and the best way to develop insight
is often to visualize data. Visualization deserves an entire lecture of
its own, but we can explore a few features of Python’s
matplotlib library here. While there is no official
plotting library, matplotlib is the de facto
standard. First, we will import the pyplot module from
matplotlib and use two of its functions to create and
display a heat map of our
data:
Episode Prerequisites
If you are continuing in the same notebook from the previous episode,
you already have a reshaped_data variable and have imported
numpy.
For ease, let’s rename the variable:
If you are starting a new notebook at this point, you need the following three lines:
PYTHON
import numpy
data = numpy.loadtxt(fname='wavesmonthly.csv', delimiter=',', skiprows=1)
data = numpy.reshape(data[:,2], [37,12])…or, if you saved the reshaped data into a file
Now let’s use the Matplotlib library to plot this data. We’ll need to
import from the matplotlib.pyplot library. We can then use
the imshow function to plot this data. In some versions of
Python we’ll need to call the show function to display the
graph. This will plot a heatmap of our data.
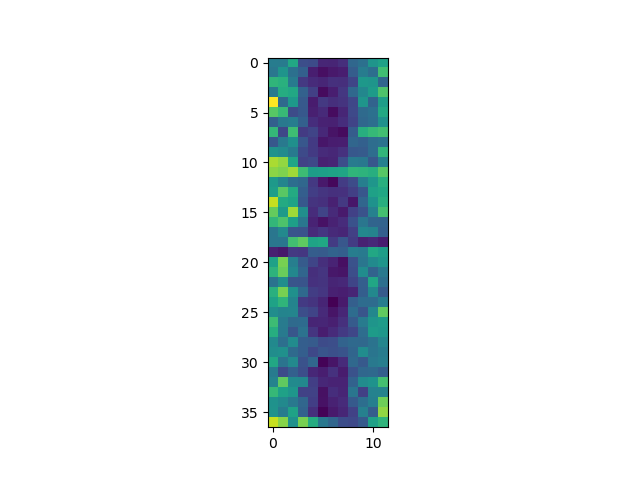
Each row in the heat map corresponds to a year in the dataset, and each column corresponds to a month. Blue pixels in this heat map represent low values, while yellow pixels represent high values. We can see low blue values in the middle (summer) months, and higher waves at the start and end of the year. This demonstrates that there is a seasonal cycle present. With calm summers bringing lower waves, and windy winters generating big waves. There are still differences year to year, with some stormier summers and calmer winters.
Now let’s take a look at the average wave-height per month over time.
Here we’ll used the plot function to plot a line graph.
PYTHON
ave_waveheight = numpy.mean(data, axis=0)
ave_plot = matplotlib.pyplot.plot(ave_waveheight)
matplotlib.pyplot.show()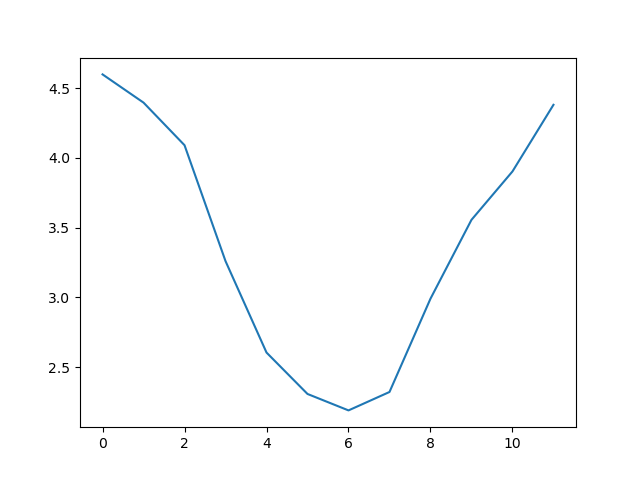
This is a good way to smooth out variability, and see what is called a ‘climatology’, representing the long-term wave climate over several years or decades.
Here, we have put the average wave heights per month across all years
in the array ave_waveheight, then asked
matplotlib.pyplot to create and display a line graph of
those values. The result is a smooth seasonal cycle, with a maximum in
month 0 (January) and minimum in month 6 (July). But a good data
scientist doesn’t just consider the average of a dataset, so let’s have
a look at the minimum and maximum too. It’s good practice to add axes
labels to our graphs, these can be done with the xlabel and
ylabel functions in matplolib.pyplot.
PYTHON
matplotlib.pyplot.ylabel("Max Wave Height (metres)")
matplotlib.pyplot.xlabel("Month")
max_plot = matplotlib.pyplot.plot(numpy.max(data, axis=0))
matplotlib.pyplot.show()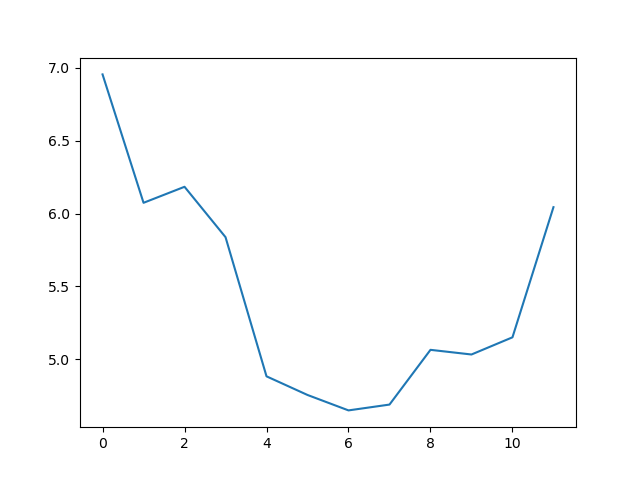
PYTHON
matplotlib.pyplot.ylabel("Min Wave Height (metres)")
matplotlib.pyplot.xlabel("Month")
min_plot = matplotlib.pyplot.plot(numpy.min(data, axis=0))
matplotlib.pyplot.show()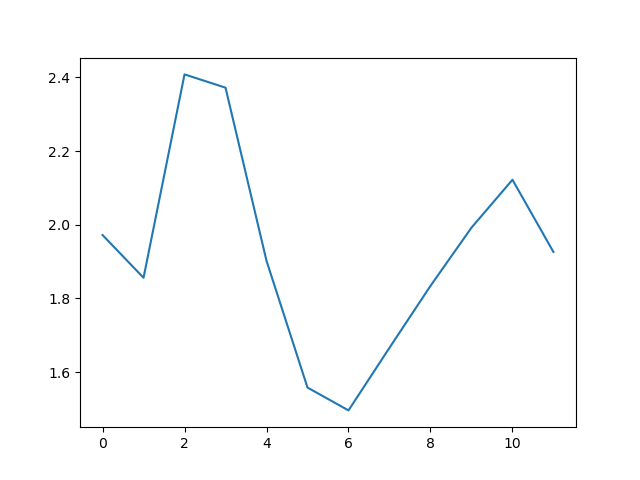
The minimum and maximum graphs show the large spread of all possible wave heights throughout the dataset. There is still a seasonal cycle, but less clear as the extremes are much less smooth. The maximum wave heights can reach a massive 7 metres, and even in the summer the maximum is 4.5m (around the height of a double decker bus!) The minimum values are more similar throughout the year, varying between 1.5 and 2.5 metres.
Plotting the data in this way, allows us to get a broad picture of the wave climate, without having to examine the numbers themselves without visualization tools.
Grouping plots
You can group similar plots in a single figure using subplots. This
script below uses a number of new commands. The function
matplotlib.pyplot.figure() creates a space into which we
will place all of our plots. The parameter figsize tells
Python how big to make this space. Each subplot is placed into the
figure using its add_subplot method. The add_subplot
method takes 3 parameters. The first denotes how many total rows of
subplots there are, the second parameter refers to the total number of
subplot columns, and the final parameter denotes which subplot your
variable is referencing (left-to-right, top-to-bottom). Each subplot is
stored in a different variable (axes1, axes2,
axes3). Once a subplot is created, the axes can be titled
using the set_xlabel() command (or
set_ylabel()), note these are different to the
xlabel and ylabel functions used by
matplot.pyplot. Let’s create three new plots, side by side,
this time showing within each of the 37 years of the dataset - notice
how we now use axis=1 in our calls to the summary statistic
functions:
PYTHON
fig = matplotlib.pyplot.figure(figsize=(10.0, 3.0))
axes1 = fig.add_subplot(1, 3, 1)
axes2 = fig.add_subplot(1, 3, 2)
axes3 = fig.add_subplot(1, 3, 3)
axes1.set_ylabel('Average')
axes1.set_xlabel('Year index')
axes1.plot(numpy.mean(data, axis=1))
axes2.set_ylabel('Max')
axes2.set_xlabel('Year index')
axes2.plot(numpy.max(data, axis=1))
axes3.set_ylabel('Min')
axes3.set_xlabel('Year index')
axes3.plot(numpy.min(data, axis=1))
fig.tight_layout()
matplotlib.pyplot.savefig('wavedata.png')
matplotlib.pyplot.show()This script tells the plotting library how large we want the figure
to be, that we’re creating three subplots, what to draw for each one,
and that we want a tight layout. (If we leave out that call to
fig.tight_layout(), the graphs will actually be squeezed
together more closely.)
The call to savefig stores the plot as a graphics file.
This can be a convenient way to store your plots for use in other
documents, web pages etc. The graphics format is automatically
determined by Matplotlib from the file name ending we specify; here PNG
from ‘wavedata.png’. Matplotlib supports many different graphics
formats, including SVG, PDF, and JPEG.
Importing libraries with shortcuts
So far we use have used the code
import matplotlib.pyplot syntax to import the
pyplot module of matplotlib. An alternative
method for importing is to use
import matplotlib.pyplot as plt. Importing
pyplot this way means that after the initial import, rather
than writing matplotlib.pyplot.plot(...), you can now write
plt.plot(...). Another common convention is to use the
shortcut import numpy as np when importing the NumPy
library. We then can write np.loadtxt(...) instead of
numpy.loadtxt(...), for example.
Some people prefer these shortcuts as it is quicker to type and
results in shorter lines of code - especially for libraries with long
names! You will frequently see Python code online using a
pyplot function with plt, or a NumPy function
with np, and it’s because they’ve used this shortcut. It
makes no difference which approach you choose to take, but you must be
consistent as if you use import matplotlib.pyplot as plt
then matplotlib.pyplot.plot(...) will not work, and you
must use plt.plot(...) instead. Because of this, when
working with other people it is important you agree on how libraries are
imported. From this point onwards this lesson uses plt to
mean matplotlib.pyplot.
Plot Scaling
Why do all of our plots stop just short of the upper end of our graph?
Because matplotlib normally sets x and y axes limits to the min and max of our data (depending on data range)
Plotting multiple graphs on one pair of axes
We can also plot more than one dataset on a single pair of axes, and Matplotlib gives us lots of control over the output. Can you plot the maximum, minimum, and mean all on the same axes, change the colour and marker used for each of the plots, and give the plot a legend?
We can call plot multiple times before we call
show, and each of those will be added to the axes. We can
also specify format options as a string (this needs to specified
straight after the data to plot), with all available options listed in
the
documentation. We also need to specify the label for
each plot, and call legend() to make the legend
visible.
An example would be
PYTHON
import matplotlib.pyplot as plt
plt.plot(numpy.max(data, axis=0), "bo", label='Maximum')
plt.plot(numpy.average(data, axis=0), "m+", label='Average')
plt.plot(numpy.min(data, axis=0), "r--", label='Minumum')
plt.legend(loc='best')
plt.show()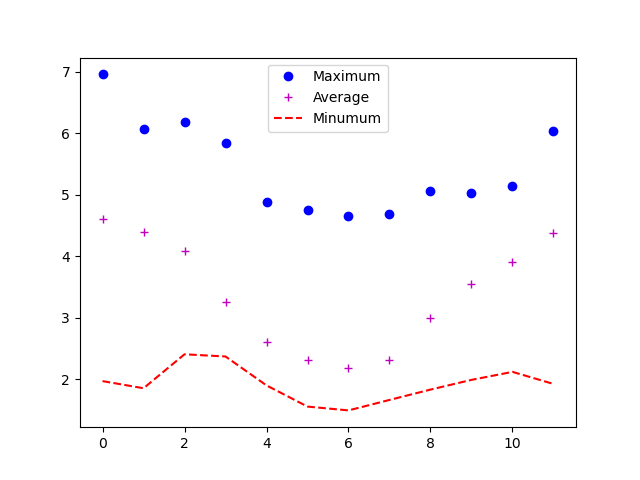
Make Your Own Plot
Create a plot showing the standard deviation (numpy.std)
of the wave data across all months.
Moving Plots Around
Modify the program to display the three plots on top of one another instead of side by side.
PYTHON
# change figsize (swap width and height)
fig = plt.figure(figsize=(3.0, 10.0))
# change add_subplot (swap first two parameters)
axes1 = fig.add_subplot(3, 1, 1)
axes2 = fig.add_subplot(3, 1, 2)
axes3 = fig.add_subplot(3, 1, 3)
axes1.set_ylabel('average')
axes1.plot(numpy.mean(data, axis=1))
axes2.set_ylabel('max')
axes2.plot(numpy.max(data, axis=1))
axes3.set_ylabel('min')
axes3.plot(numpy.min(data, axis=1))
fig.tight_layout()
plt.show()NetCDF files
What about data stored in other types of files? Scientific data is often stored in NetCDF files. We can also read these files easily with python, but we use to use a different library
We will again use data describing sea waves, but this time looking at a spatial map. This data set shows a static world map, containing data with the multi-year average wave climate. Again, hs_avg is the wave height in metres. But this time, the shape of the matrix is latitude x longitude
Using other libraries
For the rest of this lesson, we need to use a python library that isn’t included in the default installation of Anaconda. There are various ways to doing this, depending on how you opened the Jupyter Notebook:
If you’re using Anaconda Navigator: - return to the main window of
Anaconda Navigator - select “Environments” from the left-hand menu, and
then “base (root)” - Select the Not Installed filter
option to list all packages that are available in the environment’s
channels, but not installed. - Select the name of the package you want
to install. We want NetCDF4 - Click Apply
If you opened the Jupyter Notebook via the command line: - you’ll
need to close the Notebook (Ctrl+C, twice) - run the command
conda install netcdf4, and accept any prompt(s)
(y)
We can then import a netCDF file, and check to see what python thinks its type is:
OUTPUT
<class 'netCDF4._netCDF4.Dataset'>We can use Matplotlib to display this data set as a world map. The data go from -90 degrees (south pole) to +90 degrees (north pole) in the y direction. In the x-direction the data go from 0 to 360 degrees East, starting at the Greenwich meridian. The white areas are land, because we have no data there to plot.
PYTHON
plt.xlabel("Longitude")
plt.ylabel("Latitude")
plt.imshow(globaldata["hs_avg"][0], extent=[0,360,-90,90], origin='lower')
plt.show()NetCDF files can be quite complex, and normally consist of a number
of variables stored as 2D or 3D arrays.
globaldata["hs_avg"] is getting a variable called
hs_avg (average significant wave height), which is of type
netCDF4._netCDF4.Variable (the full list of variables
stored can be listed with globaldata.variables.keys()). We
can use the first element of globaldata["hs_avg"] to plot
the global map using the imshow() function. Although the
type of this element is numpy.ma.core.MaskedArray, the
imshow() function can natively use this variable type as
input.
We then need to specify that the data axes of the plot need to go
from 0 to 360 on the x-axis, and -90 to 90 on the y-axis. We also use
origin='lower' to stop the map being displayed upside down,
because we want the map being plotted from the bottom-left, rather than
the top-left which is the default for plotting matrix-type data (because
this is where [0:0] normally is).
We can also add a colour bar to help describe the figure, with a little more code:
PYTHON
fig = plt.figure(figsize=(12.0,4.0))
ax = plt.gca()
ax.set_xlabel("Longitude")
ax.set_ylabel("Latitude")
im = ax.imshow(globaldata["hs_avg"][0], extent=[0,360,-90,90], origin='lower')
cbar = fig.colorbar(im, ax=ax, location="right", pad=0.02)
plt.show()Investigate NetCDF Metadata
NetCDF files don’t just include data, they also include metadata
describing their data. If we print the globaldata object we
can see some of this data.
Print the dataset and find out the following:
- When was this dataset created?
- How many timesteps are there in the data?
- What date range does the data cover?
Accessing subsets of NetCDF data
How might we get the value of first timestep at the Northernmost and Westernmost point?
How could we access a North/South transect through the data at the 180th longitude point and first timestep? Make a graph of this data.
- “Use the
pyplotmodule from thematplotliblibrary for creating simple visualizations.”
